This function computes the phylogenetically based transcriptome evolutionary index (TEI) similar to Domazet-Loso & Tautz, 2010.
TEI(
ExpressionSet,
Phylostratum = NULL,
split = 1e+05,
showprogress = TRUE,
threads = 1
)Arguments
- ExpressionSet
expression object with rownames as GeneID (dgCMatrix) or standard PhyloExpressionSet object.
- Phylostratum
a named vector representing phylostratum per GeneID with names as GeneID (not used if Expression is PhyloExpressionSet).
- split
specify number of columns to split
- showprogress
boolean if progressbar should be shown
- threads
specify number of threads
Value
a numeric vector containing the TEI values for all given cells or developmental stages.
Details
The TEI measure represents the weighted arithmetic mean (expression levels as weights for the phylostratum value) over all evolutionary age categories denoted as phylostra.
$$TEI_s = \sum (e_is * ps_i) / \sum e_is$$
where TEI_s denotes the TEI value in developmental stage s, e_is denotes the gene expression level of gene i in stage s, and ps_i denotes the corresponding phylostratum of gene i, \(i = 1,...,N\) and N = total number of genes.
References
Domazet-Loso T. and Tautz D. (2010). A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature (468): 815-818.
Quint M et al. (2012). A transcriptomic hourglass in plant embryogenesis. Nature (490): 98-101.
Drost HG et al. (2015) Mol Biol Evol. 32 (5): 1221-1231 doi:10.1093/molbev/msv012
Examples
## get Seurat object
celegans<-readRDS(file=system.file("extdata",
"celegans.embryo.SeuratData.rds", package="scTEI")
)
## load Caenorhabditis elegans gene age estimation
celegans_ps<-readr::read_tsv(
file=system.file("extdata",
"Sun2021_Orthomap.tsv", package="scTEI")
)
#> Rows: 20040 Columns: 2
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: "\t"
#> chr (1): GeneID
#> dbl (1): Phylostratum
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
## define Phylostratum
ps_vec<-setNames(
as.numeric(celegans_ps$Phylostratum),
celegans_ps$GeneID
)
## add TEI values
celegans@meta.data["TEI"]<-TEI(
ExpressionSet=Seurat::GetAssayData(celegans, assay="RNA", layer="counts"),
Phylostratum=ps_vec
)
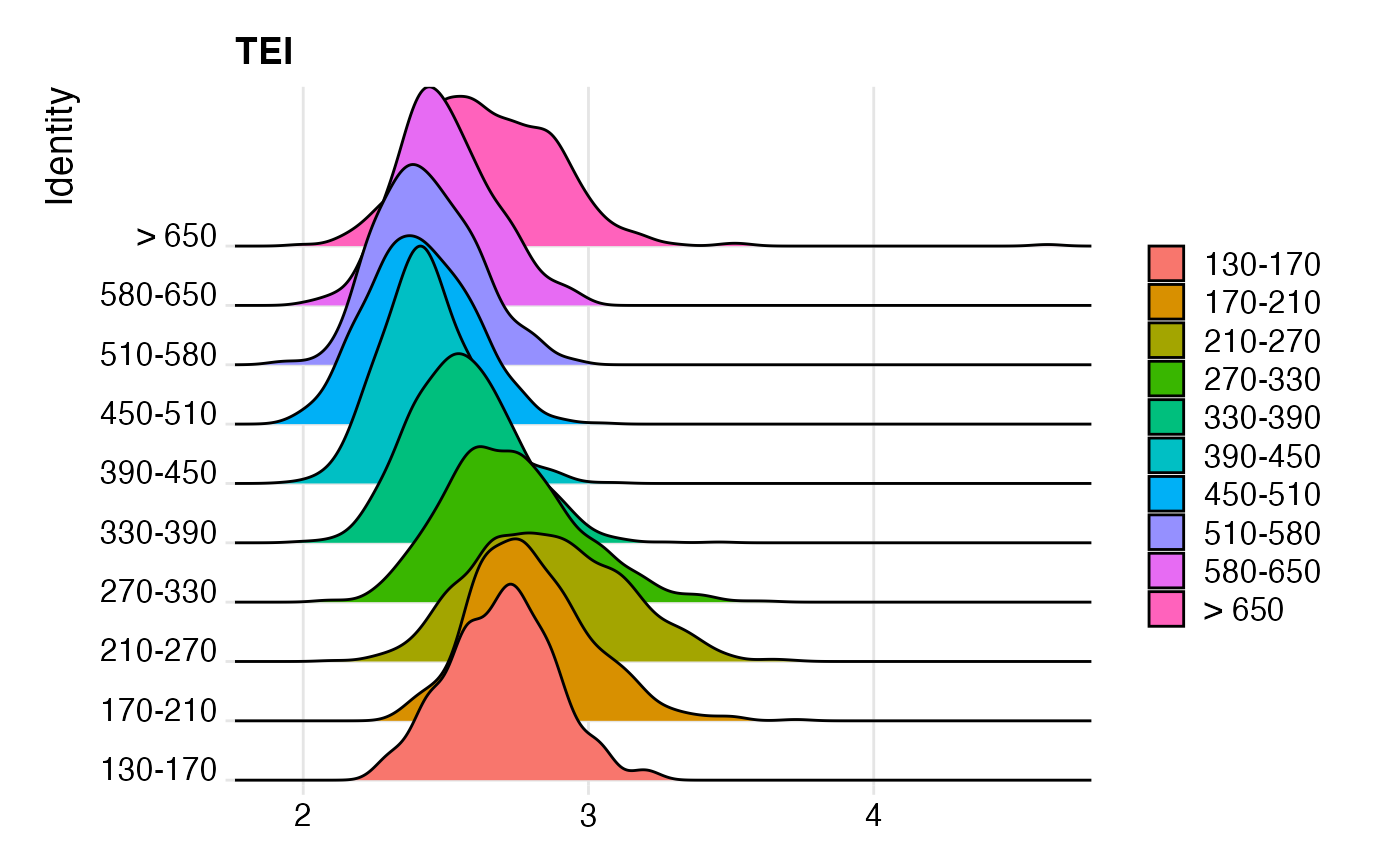
## make RidgePlot
Seurat::Idents(celegans)<-"embryo.time.bin"
p1<-Seurat::RidgePlot(
object=celegans,
features="TEI"
)
#> Warning: The `slot` argument of `FetchData()` is deprecated as of SeuratObject 5.0.0.
#> ℹ Please use the `layer` argument instead.
#> ℹ The deprecated feature was likely used in the Seurat package.
#> Please report the issue at <https://github.com/satijalab/seurat/issues>.
#> Warning: `PackageCheck()` was deprecated in SeuratObject 5.0.0.
#> ℹ Please use `rlang::check_installed()` instead.
#> ℹ The deprecated feature was likely used in the Seurat package.
#> Please report the issue at <https://github.com/satijalab/seurat/issues>.
p1
#> Picking joint bandwidth of 0.0512