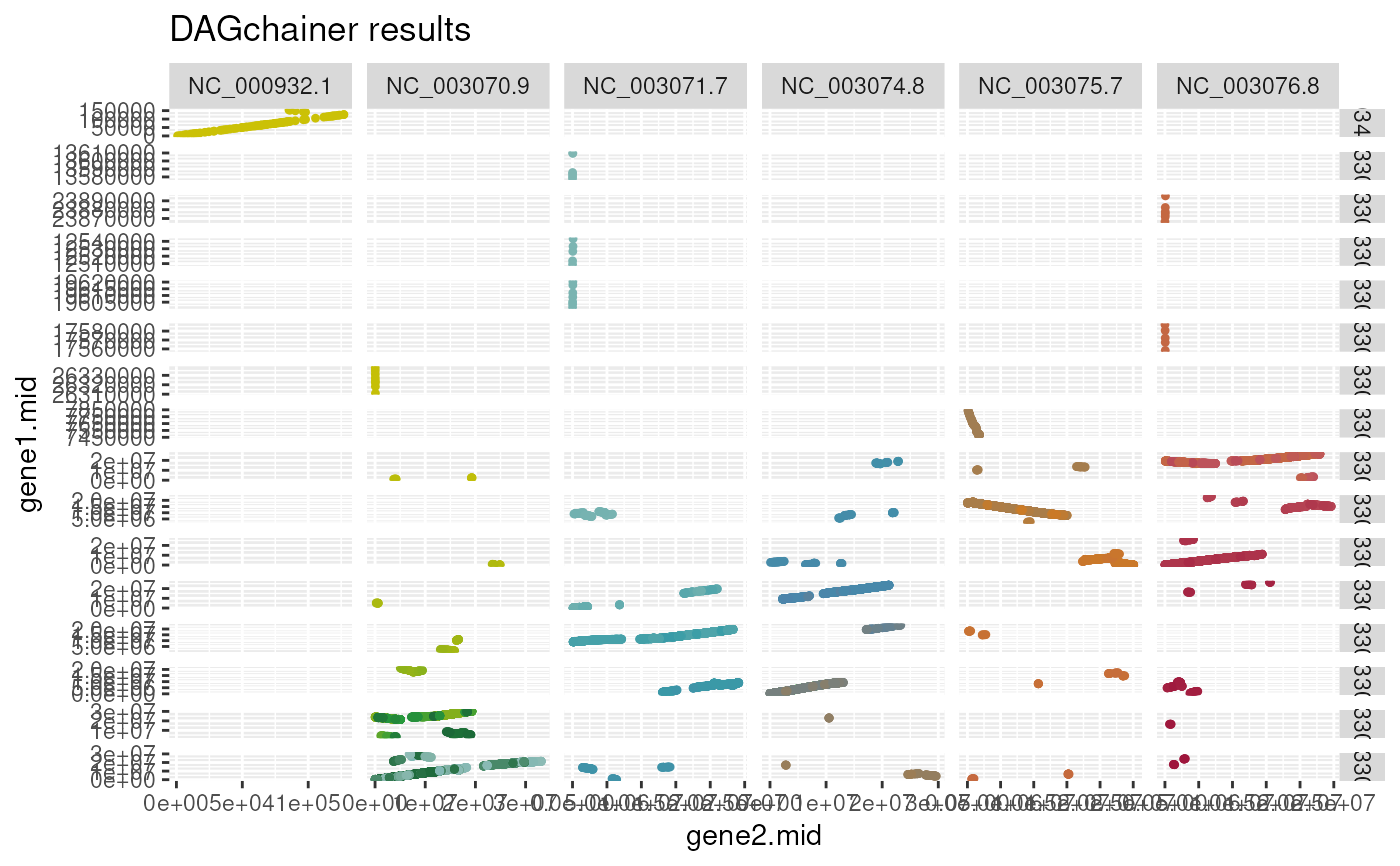
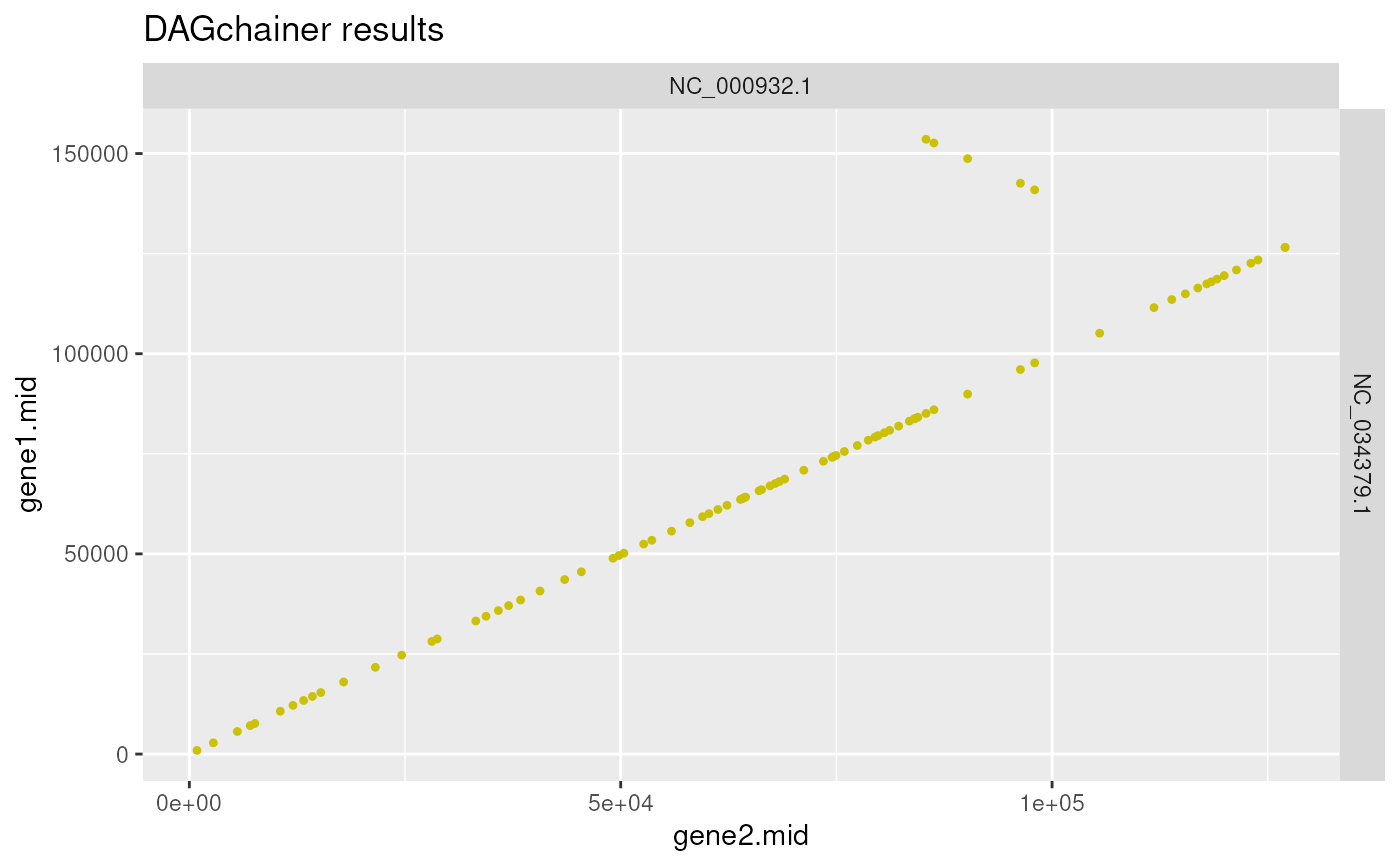
This function plots DAGchainer (http://dagchainer.sourceforge.net/) results obtained via `rbh2dagchainer()` function.
Usage
plot_dagchainer(
dag,
DotPlotTitle = "DAGchainer results",
colorBy = "none",
kaks = NULL,
ka.max = 5,
ks.max = 5,
ka.min = 0,
ks.min = 0,
select.chr = NULL
)Arguments
- dag
specify DAGchainer results as obtained via `rbh2dagchainer()` [mandatory]
- DotPlotTitle
specify DotPlot title [default: DAGchainer results]
- colorBy
specify if dagchainer groups should be colored by "Ka", "Ks", "Ka/Ks" or "none" [default: none]
- kaks
specify Ka/Ks input obtained via `rbh2kaks()` [default: NULL]
- ka.max
specify max Ka to be filtered [default: 5]
- ks.max
specify max Ks to be filtered [default: 5]
- ka.min
specify min Ka to be filtered [default: 0]
- ks.min
specify min Ks to be filtered [default: 0]
- select.chr
filter results for chromosome names [default: NULL]